Genetic risk prediction of gene expression, complex traits, and polygenic disease
Data science capstone domain of inquiry (DSC 180AB 2024-2025)
Developed by Tiffany Amariuta.
Introduction
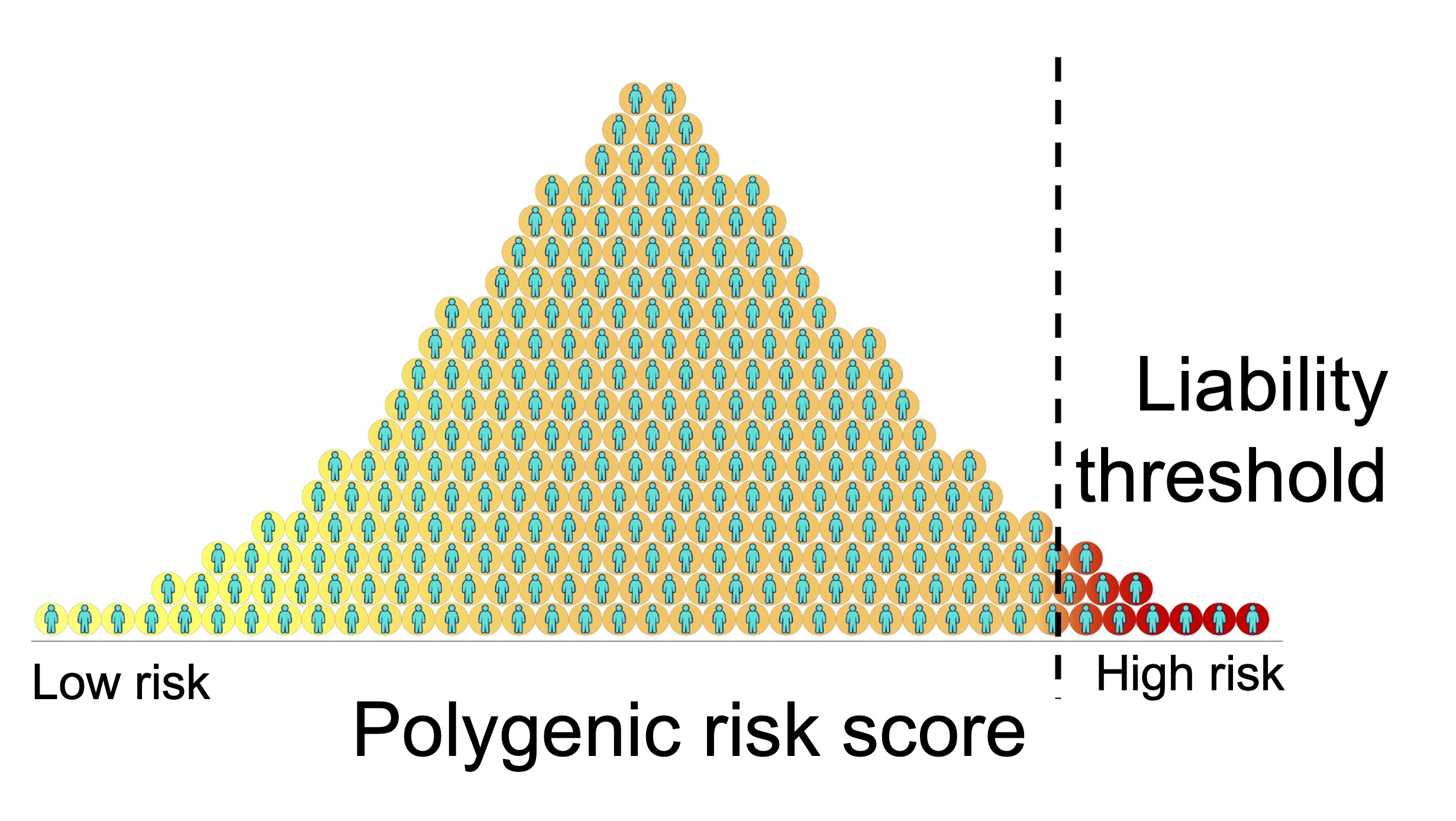
A genetic risk score (GRS) or polygenic risk score (PRS) is a weighted sum of an individual’s risk alleles across one’s genome for a particular phenotype, i.e. disease or other measurement. The weights are typically the effect sizes of the risk allele, estimated by a genome-wide association study (in the case of complex traits / polygenic diseases) or an eQTL study (in the case of gene expression). PRS have great potential to revolutionize preventive care. In theory, an individual may arrive at the clinic not knowing their genetic susceptibility to a disease, have their DNA sequenced, and learn what is their lifetime risk for the disease. There is a theoretical liability threshold of PRS at which individuals who have a PRS value lower than the threshold will not develop the disease and those with a value higher than the threshold will develop disease. For diseases with a monogenic basis, it has been shown that the same degree of disease risk can be conferred by polygenic risk alone (Khera 2018 Nature Genetics). PRS are generally useful for understanding how predictive genetics is of disease and how disperse the genetic contributions are. PRS is especially useful in understanding genetic liability when individual effects are too small to be easily detected by genome-wide association studies (Purcell 2009 Nature).
In this capstone, students will use population genetics and genomics data to assess individual risk for disease outcomes and transcriptomic measurements. Students will learn to work with genotype data from 1000Genomes and genetic association data from genome-wide association studies (GWAS) and transcriptome-wide association studies (TWAS).
In this capstone, we will be trying to answer the following questions:
- What proportion of disease-associated genetic variation is propagated through gene expression?
- How does genetic liability (high PRS) for gene expression correlate with genetic liability for polygenic disease?
- What explains the missing phenotypic variance of gene expression and polygenic disease?
To answer these questions, we will be working toward performing the following types of analysis:
- Detection of genetic effects on gene expression (eQTL analysis)
- Identification of disease-associated genes: colocalization analysis of eQTL and GWAS data
- From association to causation: fine-mapping analysis of detected eQTLs
Reading and resources
In the first part of the capstone, we will be getting familiar with human genotype and phenotype data. While human genotyping data tends to be heavily protected due to privacy concerns and reidentification of individuals, the 1000Genomes consortium is a publicly available resource that we can work with:
- [1000Genomes / Geuvadis Manuscript] note: do not go to http://www.geuvadis.org/ (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3918453/)
- 1000Genomes Genetic Data
Section Participation
Participation in the weekly discussion section is mandatory. Each week, you are responsible for doing the reading/task assigned in the schedule. Come to section prepared to ask questions about and discuss the results of these tasks.
Each week, turn in answers to the weekly questions to Gradescope. These questions are meant to focus your work for the week and help prepare you for discussion. If you have questions about your work, please ask them in section or office hours.
You are responsible for the entire weekly reading/task, even if portions are not covered in the weekly questions. The weekly tasks are the building blocks for the project proposals/assignments due at the end of the quarter.
Schedule
Office Hours
Alternating between Friday 11AM-12PM and Thursday 2-3PM, Franklin Antonio Hall, 2nd floor south-west collaboratory 2101.